
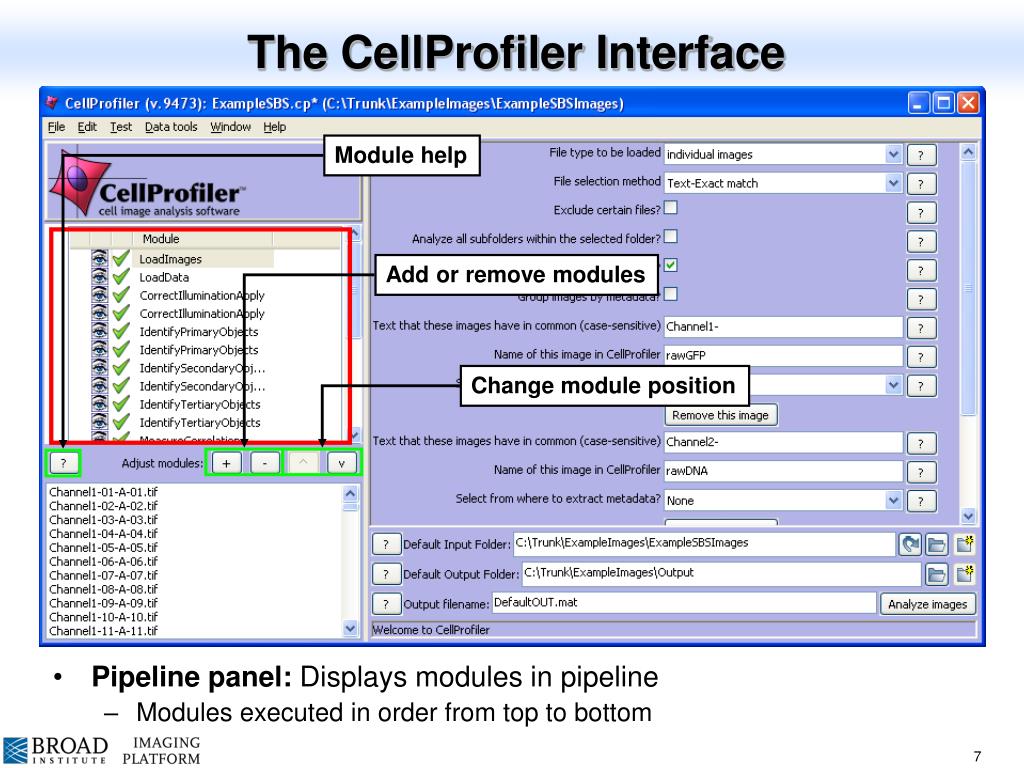
Once configured, RunCellpose should execute just like any other CellProfiler module. This allows Cellpose to operate on data from different magnifications without needing specific training. Internally, the image will be rescaled to match the object diameter which the loaded model was trained to detect. Most importantly, RunCellpose needs to know the expected diameter of the objects that you’re trying to detect. Input images are then chosen, with the option to supply a nuclear stain being available when detecting cells. The module is configured by selecting whether to detect cells, nuclei or a custom model supplied by the user. Additional help and information is available by clicking on the ‘?’ buttons next to each setting. The plugin behaves as a standard CellProfiler module with a series of settings. This may mean that CellProfiler takes some time to start up after installing the plugin for the first time. On first run Cellpose will automatically download its built-in models from the internet. The plugin module includes a testing function to check if a GPU is configured correctly. Running on a GPU is substantially faster than on a CPU, so it’s well worth trying this if your system supports it. To do so you’ll need to follow the instructions here to configure a PyTorch version for your particular machine. If you have compatible hardware, you may also want to try running Cellpose on a graphics card (GPU). The plugin should load in and be available next time you start CellProfiler. Once that’s done, drop the RunCellpose plugin script into the ‘Plugins’ folder specified in your CellProfiler preferences.
#Cellprofiler smooth install#
On the resulting Python environment you’ll need to run `pip install cellpose` to correctly configure cellpose and the required dependencies.
#Cellprofiler smooth code#
Installationīecause Cellpose requires the PyTorch deep learning library, you’ll need to be running CellProfiler from the source code (Windows, macOS) instead of a pre-packaged build. 3D object detection can be challenging with CellProfiler’s core modules, so using Cellpose can provide good results in a fraction of the processing time. This module can also be used with 3D datasets, though we recommend running on a GPU (see below) if doing so. Cellpose excels in detecting ‘typical’ cells and nuclei which can be found in many different experiments, without the need for fine-tuning of detection parameters. Functionally the RunCellpose plugin serves as an alternative to the native IdentifyPrimaryObjects and IdentifySecondaryObjects modules. You can use the inbuilt models or even provide one you’ve trained yourself.
#Cellprofiler smooth software#
The RunCellpose plugin provides a wrapper around the Cellpose package to allow you to call this software directly within a CellProfiler pipeline. Cellpose has its own user interface for training and running networks, which allows models to be customised for specific datasets. This software can provide an accessible means of detecting objects without prior knowledge of image processing strategies. This package is supplied with several pre-trained models geared towards detection of nuclei or whole cells. Today we’re releasing the RunCellpose plugin for CellProfiler 4! This plugin is designed to allow you to use the popular Cellpose segmentation algorithm to generate object sets within a CellProfiler pipeline.Ĭellpose uses a neural network followed by post-processing steps to detect and segment objects in an image.


 0 kommentar(er)
0 kommentar(er)
